Last month, we launched the Repeat Visitor Series to celebrate - and show appreciation to - the scientists who participate in multiple meetings and/or courses in a short period of time. Suma Shetty debuted the Series and the next participant to be featured is Luke Zappia, who attended back-to-back meetings on Genome Informatics (November 1-4) and Single Cell Analyses (November 8-11). Luke had never been to Cold Spring Harbor Laboratory but made the most of his inaugural trip.
Luke is a graduate student at The University of Melbourne (Australia) and is based in Alicia Oshlack's lab in the Murdoch Children's Research Institute (MCRI). The PhD candidate is working on methods for analyzing single-cell RNA sequencing data, which is a technology that measures the activity of genes in individual cells. We chatted with him on why - besides great timing - he attended both Genome Informatics and Single Cell Analyses, and what his experience was like at the two meetings.
Was there something specific about the Genome Informatics meeting that compelled you to attend?
Members of my lab have attended past Genome Informatics meetings and we believe it to be one of the premier bioinformatics conferences. We have found it to be a great opportunity to see excellent talks and interact with people from around the world who are in the field.
How about for the Single Cell Analyses meeting?
I work with single-cell data and since the Single Cell Analyses meeting took place the week after Genome Informatics, it was a good opportunity to make the most of my trip from Australia.
Let's talk key takeaways: what was your key takeaway from Genome Informatics?
Bioinformatics is still a young field and we continue to push the boundaries of what is possible. And whenever I attend a bioinformatics meeting, I am always impressed by the wide range of problems people are tackling and the level of detail they put into their work.
What was your key takeaway from Single Cell Analyses?
People have developed a range of ways for measuring different things in individual cells beyond the common technologies I work with.
Do you have a takeaway that’s applicable to both meetings?
We are lucky to be working at a time when we are seeing a revolution in how biology is studied, leading to many exciting new discoveries and developments.
How about similarities - did you notice any between the two meetings?
Outside of both meetings having a range of interesting topics, there weren't too many similarities between them because they covered different content. As a result, each meeting attracted a distinct audience.
Did you present at either meeting?
I presented a talk at Genome Informatics during the Transcriptomics, Alternative Splicing, Gene Predictions Session, and a poster at Single Cell Analyses titled "Simulation and analysis tools for single-cell RNA sequencing data."
Thinking back on the scientific meetings and conferences you have attended, what do you like most about CSHL meetings?
The set-up of a CSHL meeting is helpful in generating opportunities for discussions. Everything is located on campus so I was able to just focus on the meeting and speak with other attendees. And the days are long at a CSHL meeting!
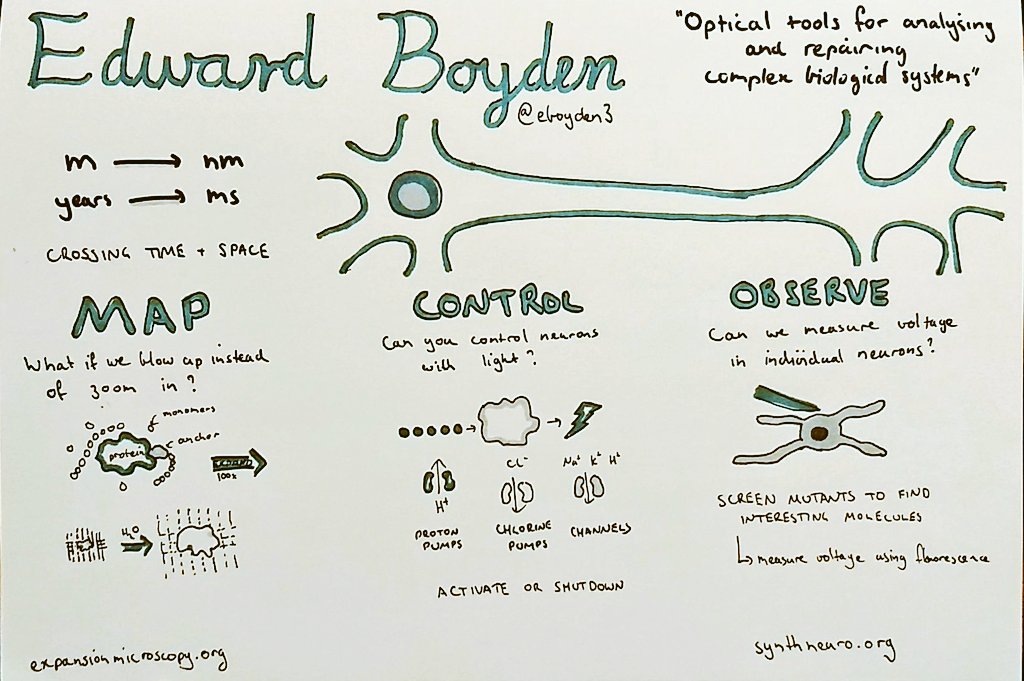
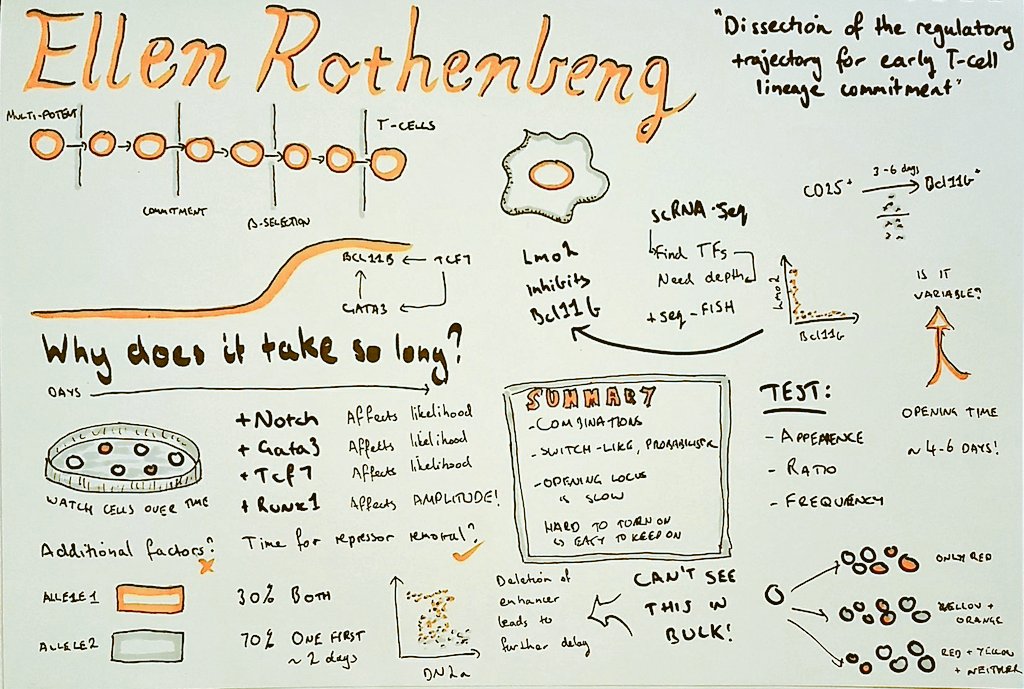
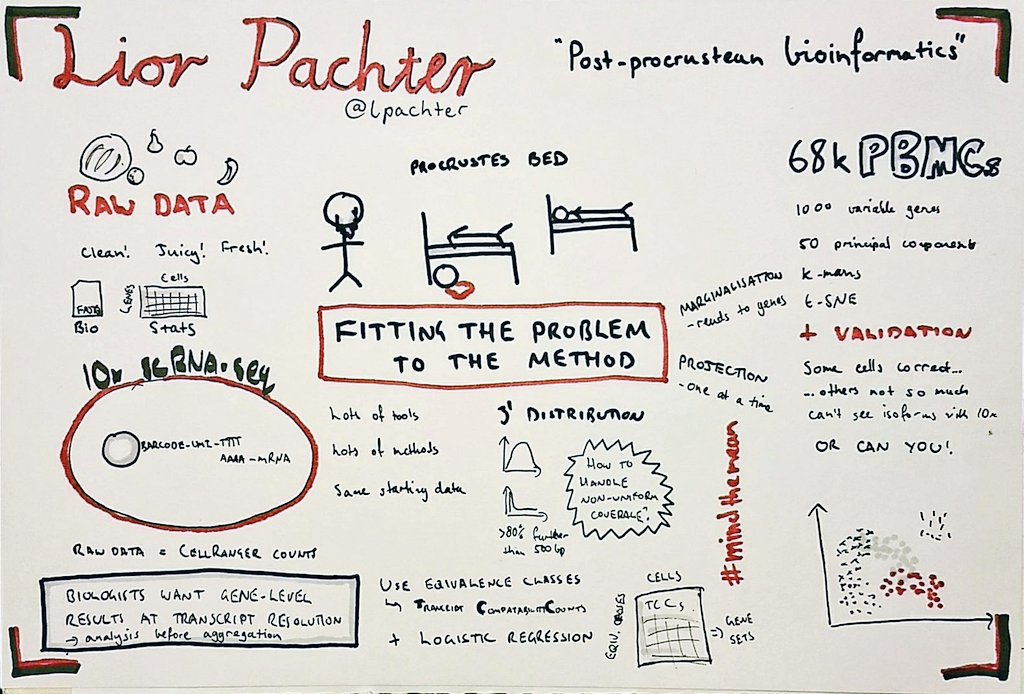

We noticed that you sketch your notes. What got you started in sketching talks?
I have been doing it for the last four years or so. I saw some examples somewhere and thought it looked like an interesting idea. Mike Rohde is generally considered responsible for populating the idea.
Can you walk us through your process of transcribing a talk into a drawing?
I usually only sketch for the longer talks (> 30 mins) as I find it hard to pick out enough things to fill a page from a shorter talk. There are lots of different ways of doing it and some people do a really good job of shorter talks such as Alex Cagan. Personally, I try to prepare before the talk by adding the speakers name, title etc. Once the talk starts I watch out for key points they make or interesting figures, which usually form the main part of the sketch. Also, I draw on paper, although it would be interesting to try a tablet at some stage.
Thank you to Luke for taking the time to chat with us at the 2017 Genome Informatics and 2017 Single Cell Analyses meetings and his sketch note process. Both meetings will return to the Laboratory in 2019. If you’re looking for a meeting in the years that Genome Informatics is not at CSHL, the Biological Data Science meeting is a great alternative. Also, we offer an annual Single Cell Analysis course during the summer.

