Meet Sam Nicholls of the University of Birmingham (United Kingdom). The postdoctoral fellow in Nick Loman’s group is currently at CSHL participating in his first meeting: Genome Informatics. In addition to giving a talk, titled "Detecting microbial transmission and engraftment after faecal microbiota transplants with long-read metagenomics and reticulatus" during the Microbial and Metagenomics session, Sam’s work was featured as the front cover of the meeting’s abstract book. He has also been taking advantage of the chance to “meet in person a bunch of familiar Twitter avatars that [he’s] been messaging over the past year or so.”
What are your research interests? What are you working on?
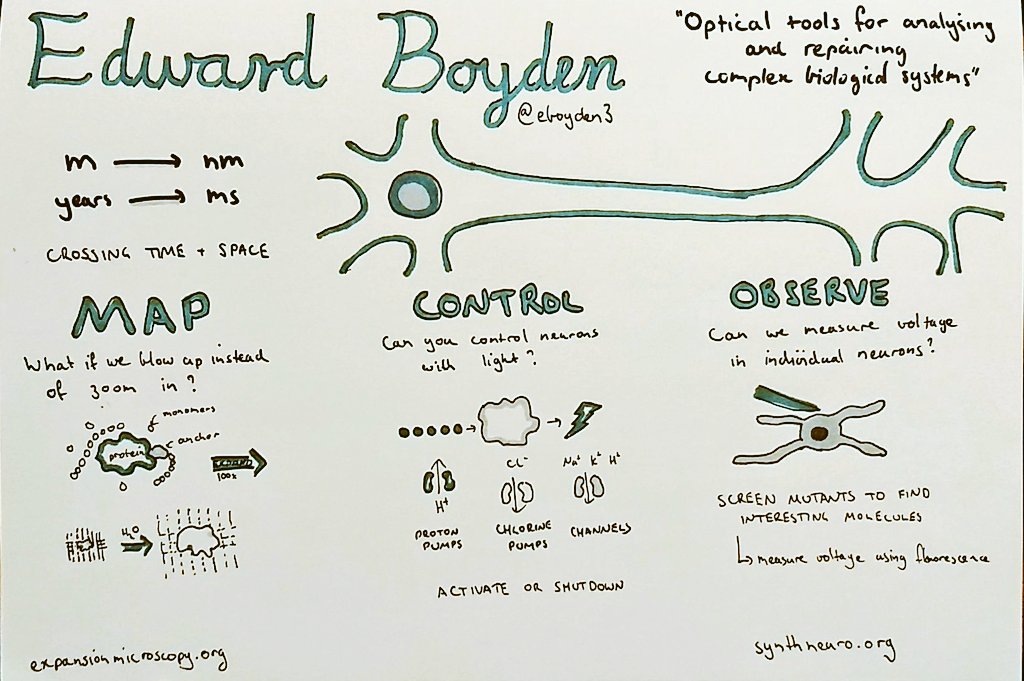
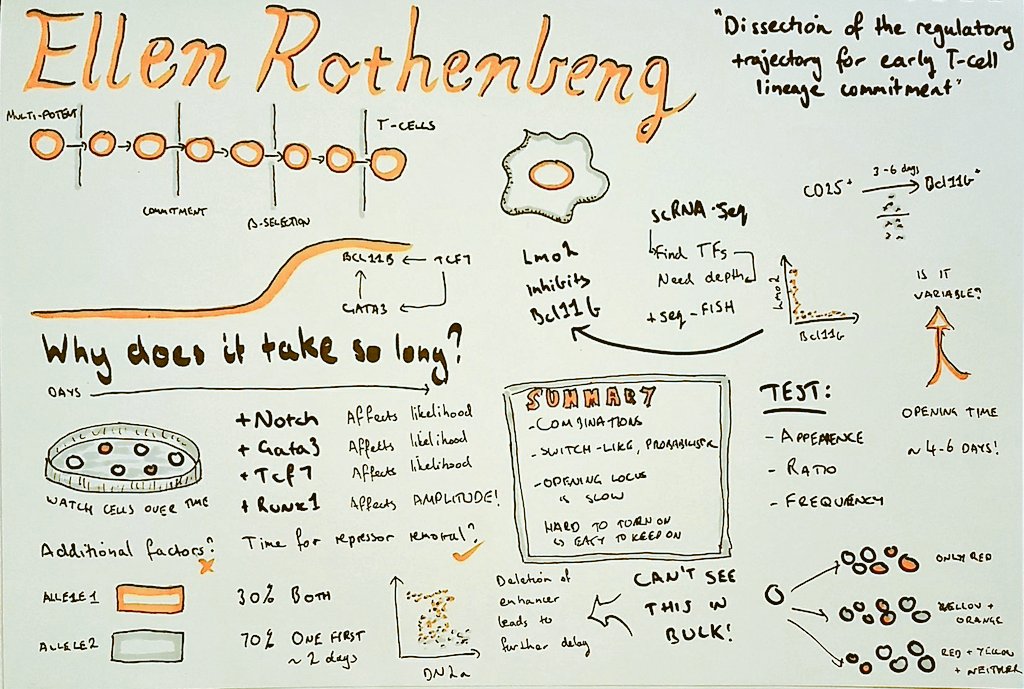
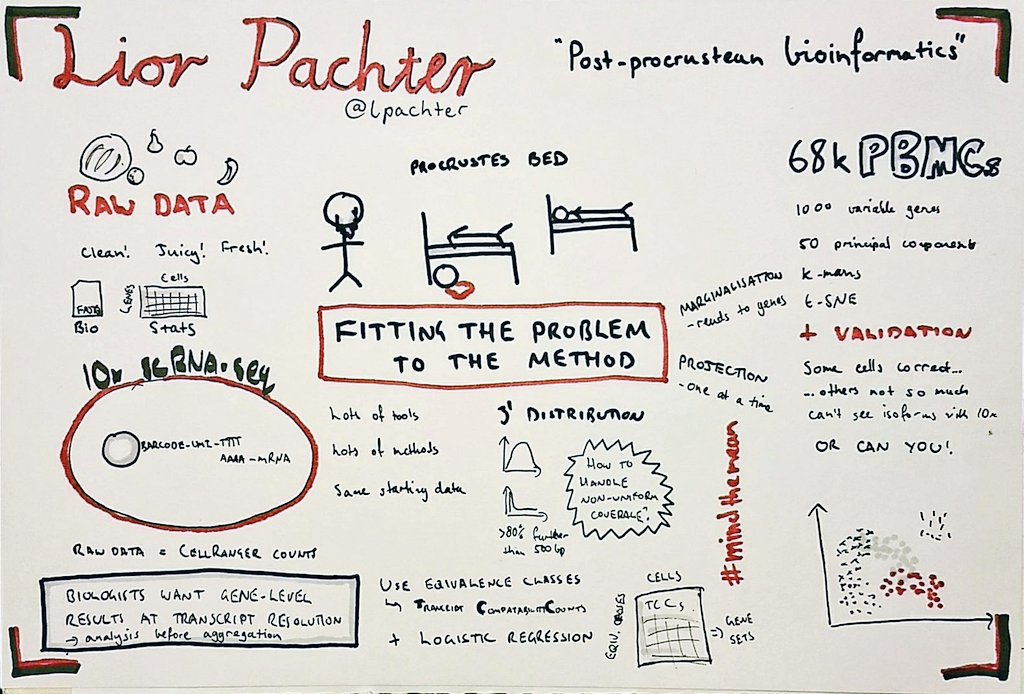
I'm currently working on computational methods to analyse clinical microbiome samples. We're interested in determining whether we can identify and characterise species and strains that are transferred from donors to recipients in faecal microbiota transplants.
How did you decided to make this to focus of your research?
This project is a natural follow-on from my PhD, which I finished a little over a year ago. My PhD was concerned with recovering haplotypes for particular genes of interest from the rumen (stomach) of a cow. We were interested in cataloguing the haplotypes of different hydrolases used to break down natural grasses in the stomach, with a view to investigate these for their potential use in the manufacture of biofuels. Now, I'm applying these skills and my experience to catalogue the genomes (and their haplotypes) that are transplanted from the gut of a healthy donor to the gut of an unwell recipient via faecal microbiota transplant, to characterise what it is about the procedure that makes it work as a successful therapy.
As part of my PhD, I created a data structure (called Hansel) for holding all possible pairwise observations of SNP variants on a set of short sequencing reads; and an algorithm (called Gretel) to use this matrix as crumbs for navigating a graph-like structure representing all possible ordered combinations of SNPs that make up the haplotypes for a region of interest. Although we didn't get so far as to test the recovered haplotypes for biofuel capabilities, as part of my final year I was able to put on a white coat and get in the lab to gain some wet lab experience of my own. I think it's important for bioinformaticians to know where their data comes from!
How did your scientific journey begin?
In the second year of my Computer Science and Statistics undergraduate degree with no plans for summer, I recall an e-mail being sent around the Computer Science Department at Aberystwyth University (United Kingdom) looking for someone who had knowledge of Perl, tree algorithms, and proteins for a placement at the Wellcome Trust Sanger Institute. Undeterred by ticking none of those boxes, I applied. The trip to Cambridge was awfully long from Aberystwyth and I didn't feel like the interview had gone particularly well. Good practice at least, I thought, but a few days later I was surprised to find that I'd been offered the summer job! On my first day, I remember someone dropping off a bunch of genetics textbooks to my desk and saying that I'd have to pick up a lot of new concepts quickly, which was a little intimidating, but I was keen to learn something completely new.
Ultimately that short placement had more of an effect on me than I had expected. I was not really aware that my skills in computing could be applied to interesting, difficult real-world problems like those we find in biology. It was nice to be writing code that wasn't just a website, or an app, or something to sell. I liked that I could tell people about my work and even post it online as it was being written, something that was definitely not part of the ethos of my previous experience in industry.
That placement led me to change my modules in my final year and to go on and pursue a PhD in bioinformatics, rather than returning to industry.
Was there something specific about the Genome Informatics meeting that drew you to attend?
As a bioinformatician, I always find myself a little lost at conferences that focus heavily on biology. I'm excited to be at Genome Informatics as this finally feels like a conference where everything makes sense to me! I was also happy at the prospect of finally meeting a bioinformatics hero of mine: Heng Li.
What is your key takeaway from the Meeting?
A takeaway in the literal sense: I'll be flying home this weekend with one of the Genome Informatics 2019 meeting posters, which features artwork that I submitted! The picture is an assembly graph from one of our complex clinical samples. It's a wonder to look at, but actually a rather terrible assembly...
What and/or how will you apply what you’ve picked up from the Meeting to your work?
Yesterday's morning session on sequencing algorithms, variant calling and genome assembly was a highlight for me so far. I'm quite interested in exploring some of the techniques for genome sketching that were discussed, and how we can apply these to our own clinical metagenomic data sets.
If someone curious in attending this meeting asked you for feedback or advice on it, what would you tell him/her?
I'm not sure how I feel about coffee at 9 in the evening, but this meeting has been full of excellent presentations that I'm going to have to go and read up more about. I'd definitely recommend Genome Informatics if you want to know what the cutting edge techniques and hot topics of bioinformatics are right now.
What do you like most about your time at CSHL?
I grew up in Swansea, Wales and moved to Aberystwyth for eight years to study my undergraduate and PhD; so I've spent most of my life near the sea. I enjoy the hustle and bustle of the city of Birmingham but I do miss the seaside. I've been enjoying walks around the campus and the views of the water. It's very calm and picturesque here. Thanks for having me!
Thank you to Sam for being this week's featured visitor. To meet other featured scientists - and discover the wide range of science that takes part in a CSHL meeting or course - go here.