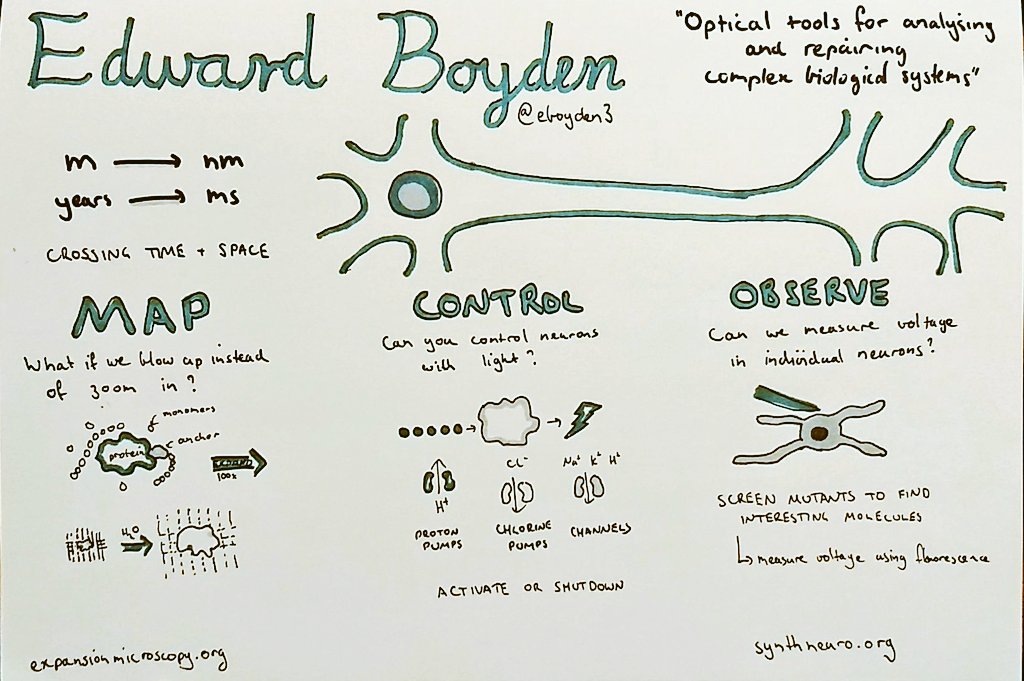
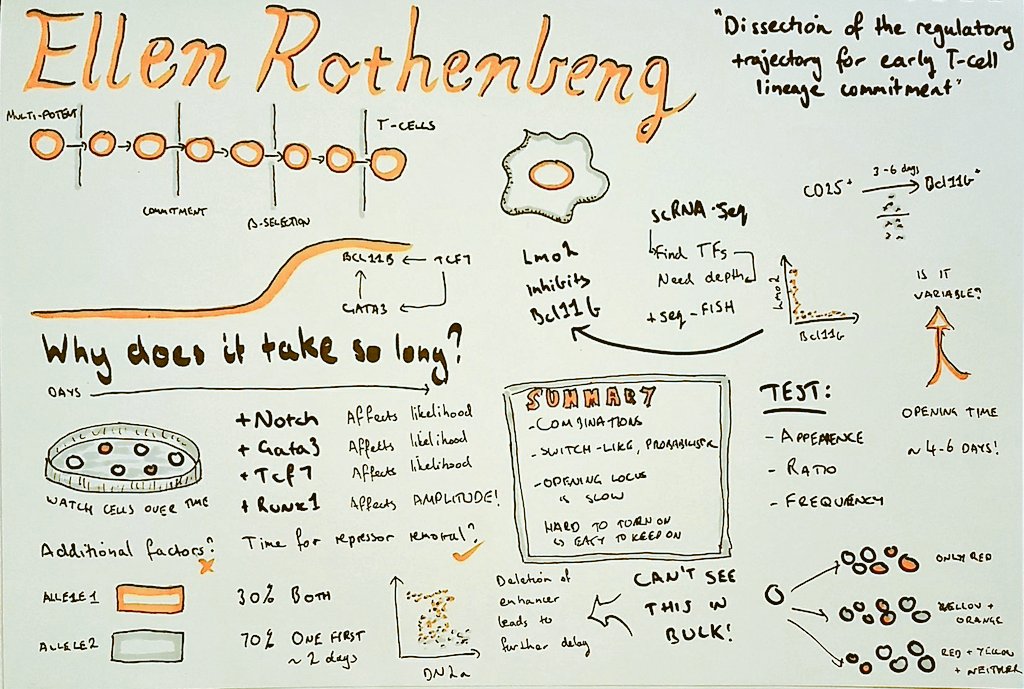
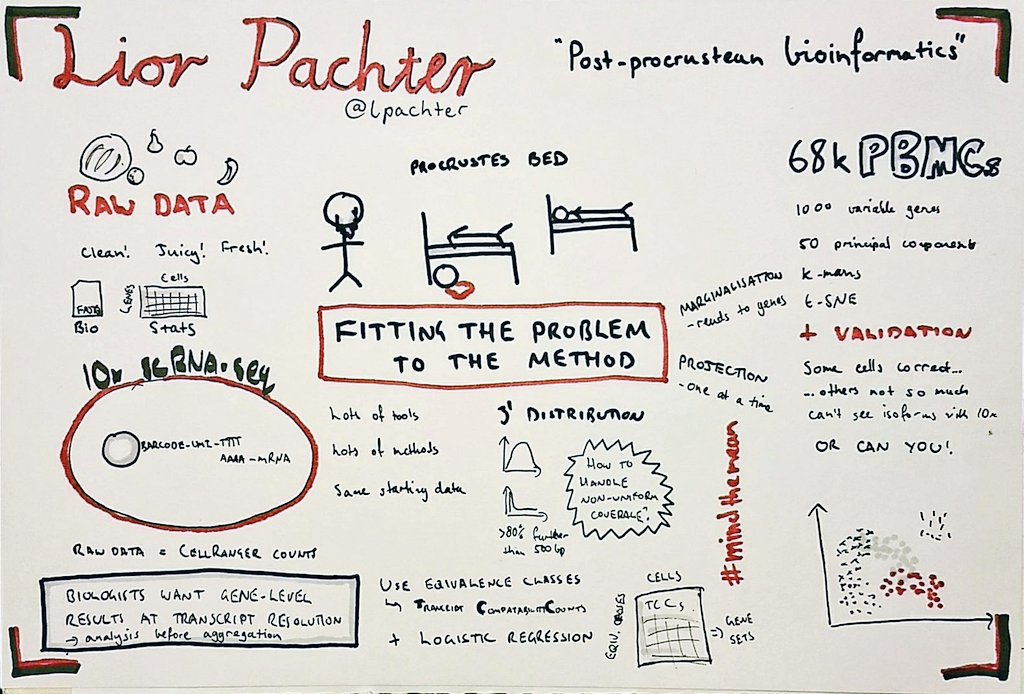
Meet Deeptiman “Deep” Chatterjee of Tulane University School of Medicine. Deep is a fifth-year Ph.D. candidate in Dr. Wu-Min Deng’s lab taking part in Single Cell Analyses. This is his first meeting at CSHL and he came into it with a bang: Deep presented a flash talk introducing his poster entitled “Constructing a Comprehensive Transcriptomic Atlas of Developing Adult Drosophila Ovary and Oogenesis”. His approach from a biological perspective without much computational made his poster different than that of the other presenters. But as they “all are working towards the same goal, [Deep was still able to have] a great experience to learn from others and get fresh feedback and good comments!”
What are your research interests? What are you working on?
My research interests lie in bridging developmental biology with that of tumor biology. I generally use classical genetics, confocal microscopy and single cell sequencing techniques in the fruit fly model system to address specific questions. In my current research, I’m trying to understand what makes specific groups of cells to consistently be more susceptible to tumor formation than their neighbors in the ovarian epithelial tissue.
How did you decide to make this the focus of your research?
My mother was a cancer survivor, and she died from cancer-related complications 11 years later. Experiencing this debilitating disease from such proximity made me appreciate this disease and I wanted to study its fundamentals at its earliest stages. Fruit flies have been extensively used to study some of these basic steps, and so when I got the opportunity to work in Dr. Wu-Min Deng’s lab, I knew I had to take it.
How did your scientific journey begin?
We had a retired biology teacher in middle school back in India, who would still teach us because he simply loved teaching. He told us to break down every complex concept into its simplest parts. I still use that logic. I also was inspired by my biology teacher in high school, who singlehandedly had set up a simple biotechnology lab for only 6 students, teaching us to standardize some of the most common protocols used in biological labs. That hands-on experience set me up for an early career in science. I went on to do an MS in yeast genetics, and I switched fields for my Ph.D.
Was there something specific about the Single Cell Analyses meeting that drew you to attend?
The methodologies used in our lab have always been wet-lab benchwork. But we have never shied away pursuing new approaches. We have recently been involved in a lot of single cell transcriptomic data generation to answer relevant questions raised in many projects in the lab. I have learnt to process and analyze these datasets in the last year by myself but was limited by my limited knowledge of the field. This being my first single cell analysis meeting of any kind, I wanted to gauge the current state of the field, to learn from it and to apply my biologically-relevant perspective to it. The opportunity to present my work as a poster allowed me better participation in a field that I’m not so familiar with.
What is your key takeaway from the Meeting?
Single cell analysis is a rapidly evolving field, and to come to meetings such as these, even for participants out of this field, is a massive learning opportunity. It also makes one appreciate the importance of interdisciplinary sciences, connecting distant fields.
What and/or how will you apply what you’ve picked up from the Meeting to your work?
I particularly liked the entire session on spatial transcriptomics, which seems most relevant to my field of research and is also really cool! I also really enjoyed the talks on RNA velocity and vector fields and would love to integrate them into my research if it applies.
If someone curious in attending this meeting asked you for feedback or advice on it, what would you tell him/her?
Definitely attend! Even if it seems irrelevant to your field, it showcases really cool science and due to the structural and functional nature of the applications of single cell techniques, it can always be applied somewhere. It’s also a great learning opportunity from some of the pioneers in the field!
Also, definitely go to the bar at the end of the day. Some of the best scientific discussions transpired there.
What do you like most about your time at CSHL?
CSHL definitely loves to feed you. Thanks for having me!
Thank you to Deep for being this week's featured visitor. To meet other featured scientists - and discover the wide range of science that takes part in a CSHL meeting or course - go here.